|
"Essentially, all models are wrong, but some are useful." George E. P. Box.
The MozAtlas orthology framework is similar to, but less restrictive, than other genome-wide orthology pipelines currently available
, and includes a set of outgroup species specifically selected to represent the Insecta.
Gene-family relationships were identified using all-vs-all Blast (Blossum 42 matrix) searches and Inparanoid clustering, with the
longest available translations for annotated proteins. Best reciprocal hits between species were grouped together
via single linkage clustering and the sequences aligned with MUSCLE. Tree topologies were subsequently reconstructed
with both dS (synonymous substitution rate), dN (nonsynonymous substitution rate), nucleotide and protein distance
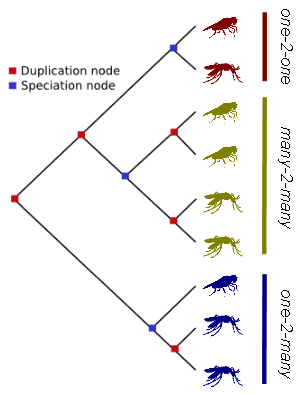
measures using TreeBest (NJ and PHYML). Orthology classifications were finally described as one-to-one, one-to-many and many-to-many gene
relationships, while gene paralogues were characterized on the basis of when divergence occurs.
|